
Summer School and Workshop 2019: Parallel Computing in Molecular Sciences
Summer School The parallel computing in molecular sciences summer school is focused on educating and training a new generation of graduate students and postdocs in molecular sciences in advanced high-performance parallel computing skills on the computing architectures of today and tomorrow, exposing the next generation of science leaders to the state-of-the-art high-performance scientific computing on […]
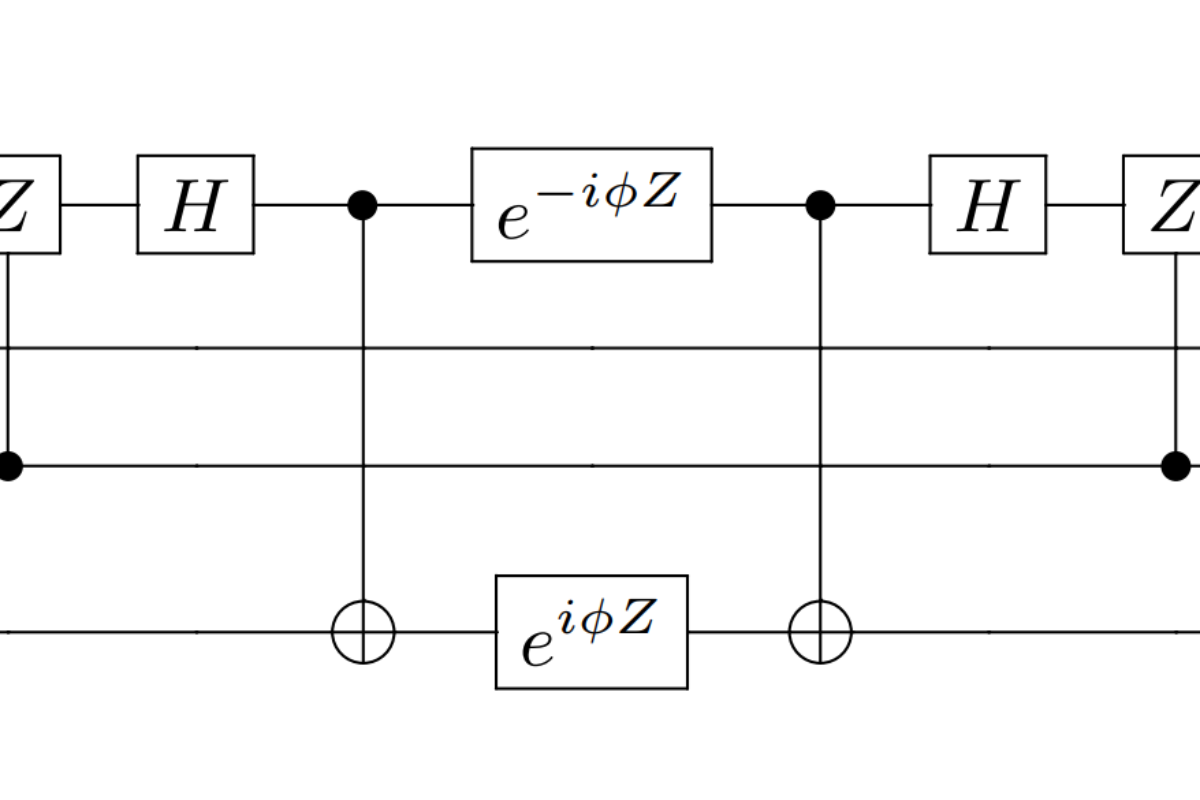
Enabling Quantum Leap: Workshop on quantum algorithms for quantum chemistry and materials
Dates: 22-24 January 2019Location: Hilton Embassy Suites in Old Town Alexandria, 1900 Diagonal Rd, Alexandria, Virginia As we begin to reach the limits of classical computers in terms of circuit size, quantum computing has emerged as a technology that has captured the imagination of the scientific world and the public as a potentially revolutionary computational advance. The […]

BioExcel/MolSSI Workshop on Workflows in Biomolecular Simulations
As part of the collaborative framework, BioExcel CoE and MolSSI are organizing a series of workshops on the application of workflow solutions for biomolecular modeling and simulations. This first event will be hosted by BioExcel in Barcelona with a planned follow-up in 2019 in the US hosted by MolSSI. Background Workflows are an increasingly important aspect of biomolecular simulation science. Over […]

Best Practices in Software Development Workshop @ SERMACS
MolSSI will hold a workshop on Best Practices in Software Development workshop at the 2018 Southeastern Regional Meeting of the American Chemical Society (SERMACS). The workshop will take place from 9 AM to 4:30 PM at the Augusta Marriott on Wednesday, October 31st. These best practices include version control with git and GitHub, unit and […]

MolSSI Workshop for SSE/SSI PIs
Organizer: Daniel Crawford Location: Kimpton Donovan Hotel, Washington, D.C. Date: April 29 2018 The MolSSI organized an afternoon workshop focused on PIs of current SSE/SSI projects that are relevant to the computational molecular sciences. The purpose of the meeting was to: inform SSE/SSI researchers of the MolSSI’s mandate and goals; to learn more about on-going […]

Summer School and Workshop Parallel Computing in Molecular Sciences
Organizers: Bert de Jong (LBNL), Robert Harrison (Stony Brook University, MolSSI), Edward Valeev (Virginia Tech), Carlos Simmerling (Stony Brook University) Location: Berkeley, CA Dates: 6-9 August 2018 The parallel computing in molecular sciences summer school is focused on educating and training a new generation of graduate students and postdocs in molecular sciences in advanced high-performance […]

MolSSI Coding Workshop @ MERCURY 2018
The MolSSI will host a coding workshop at the upcoming 2018 MERCURY Consortium conference at Furman University, July 18-19. This workshop will offer up to 50 undergraduate attendees at the conference an opportunity to learn the fundamentals of running and analyzing molecular dynamics simulations using OpenMM and MDTraj, how to use GitHub as a repository for […]

MolSSI Workshop on Tinker Modeling Software
Organizers: Pengyu Ren and Jay Ponder Location: University of Texas, Austin, Texass Dates: 2-4 June 2018 This workshop will focus on molecular modeling code development and synchronization, simulation algorithms and force field implementation, and integration between programs, as well as tutorials and applications of TINKER, TINKER-HP/OpenMM and related tools. More information is available here. A report […]

MolSSI Workshop on Python for Quantum Chemistry and Materials Simulation
Organizer: Qiming Sun Location: CalTech, Pasadena, California Dates: 29-30 June 2018 This MolSSI-sponsored workshop will focus on the development and use of Python-based open-source software stacks for quantum chemistry and materials science. A report on the workshop is available here

MolSSI Workshop on Solving or Circumventing Eigenvalue Problems in Electronic Structure Theory
Organizers: Volker Blum (Duke U.), Jianfeng Lu (Duke U.), Alvaro Vazquez-Mayagoitia (Argonne National Lab), Chao Yang (Lawrence Berkeley National Lab) Location: Richmond, Virginia Dates: 15-17 August, 2018 Workshop website: https://wordpress.elsi-interchange.org/index.php/elsi-workshop-2018/ The eigenvalue problem touches virtually every application of electronic structure theory to molecular or solid-state (materials) simulations. For small problems, traditional O(N^3) eigenvalue solver libraries are […]
